Abstract
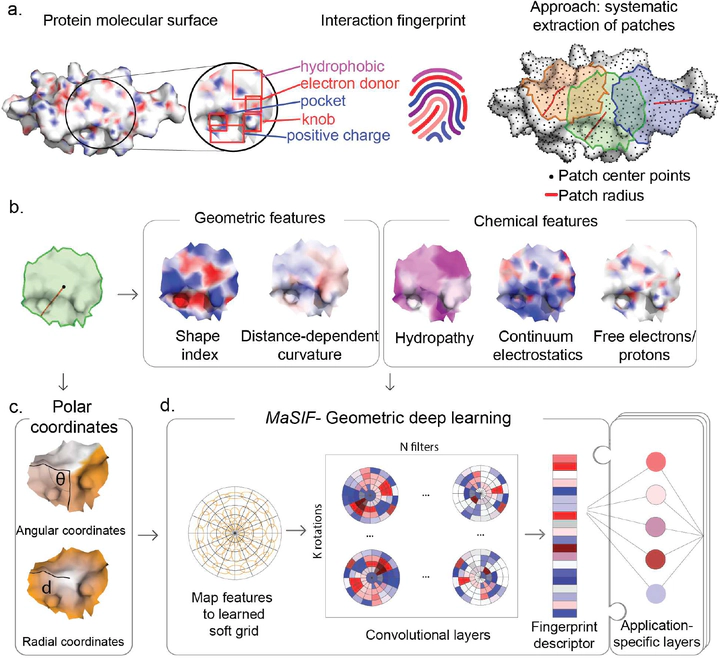
Predicting interactions between proteins and other biomolecules purely based on structure is an unsolved problem in biology. The protein molecular surface, a high-level representation of protein structure, displays the chemical and geometric features that can enter in contact with other biomolecules. This representation abstracts underlying details such as the precise arrangement of atoms and the amino acid sequence. Here we hypothesize that features in the molecular surface representation arrange in patterns, and proteins that perform similar interactions may display similar patterns. We model the discretized molecular surface as a graph and use data-driven geometric-deep learning tools to learn these patterns, and exploit them through three prediction challenges: (a) pocket similarity comparison, (b) protein-protein interaction site prediction and (c) prediction of interaction patterns between proteins based on surface patterns.